One interesting thing is that dispersion of humans from mtDNA data and Y chromosome data is similar. Yet, mtEve and yAdam span a difference of up to 100,000 years apart. From what I can tell, evolutionary theory does not predict that the migration patterns would be similar. Yet, this would logically follow from the Human Creation Model.

http://www.familytreedna.com/snps-r-us.aspx

http://news.bbc.co.uk/2/hi/science/nature/4260334.stm
https://debatingchristianity.com/forum/viewtopic.php?p=322845#p322845
nygreenguy wrote:
otseng wrote:One interesting thing is that dispersion of humans from mtDNA data and Y chromosome data is similar. Yet, mtEve and yAdam span a difference of up to 100,000 years apart. From what I can tell, evolutionary theory does not predict that the migration patterns would be similar. Yet, this would logically follow from the Human Creation Model.Thats because it doesnt really have as much to do with evolution as it does ecology. However, could you explain your point a little better (bolded) I dont really understand what your point is.
The migration patterns for both mtEve and yAdam are virtually identical. East Africa then branching out to the Middle East and rest of Africa. From Middle East branching out to Europe and Asia. From Asia to Australia and North America. From North America to South America. Now why would yAdam have a similar migration if he is tens of thousands of years later than mtEve? If man was all over the world at the time of yAdam, why would yAdam also originate in East Africa and also replace all other male lines in the same pattern at mtEve? The more parsimonious explanation was that the migrations of mtEve and yAdam happened at the same time, not separated by tens of thousands of years.
https://debatingchristianity.com/forum/viewtopic.php?p=324356#p324356
GrumpyMrGruff wrote:
otseng wrote:
GrumpyMrGruff wrote: For a good description of how drift affects mtDNA and the y-chromosome, I recommend reading this blog post.[1]I read through the blog post. But still fail to see how it explains things. Could you cite the parts where you feel answers the issue?
I’m referring to the section beginning “If we treat genes as individual organisms, we can create a “gene genealogy” – a gene family tree – for individual genes. Here’s an example I just made. … “ The blogger’s model shows in practice what others in this thread have been saying in words: Probabilistic birth-death processes (like those within animal populations) can cause an arbitrary past individual to become the MRCA for a particular gene (or mtDNA or y-chromosome) without requiring the kind of extreme post-flood bottleneck you think this implies.
However, the question is what can explain that all other female lines disappeared? In terms of the probability that only one female is the progenitor of all, I’m going to ask Zeeby in my next post about that.
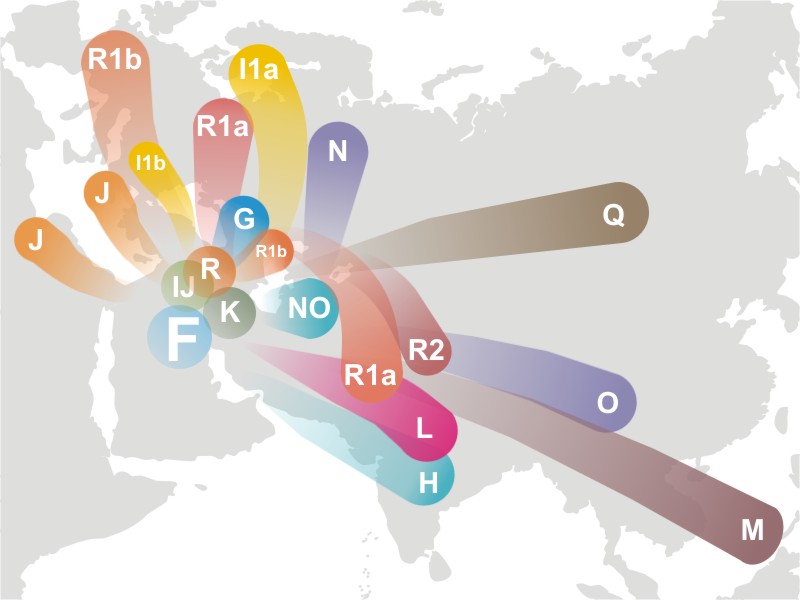
Phylogeny of y-chromosomal variants recovers the same African deep ancestry found in mtDNA lineages. Even in your flood model there is only one surviving male progenitor. If you accept the logic of this phylogeny, then you are admitting that your model is at variance with the data.
The top of the Y-chromosome mapping is the A and BT haplogroups.
http://en.wikipedia.org/wiki/Human_Y-ch … aplogroups
Here is another diagram:

https://www.sanger.ac.uk/Teams/Team19/chr-y.shtml
Haplogroup A is not limited to Africa.
Haplogroup A is found mainly in the Southern Nile region and Southern Africa. However, at lower frequencies, M91 is found in many areas of Africa, including Morocco, Egypt, and Cameroon. Outside of Africa, it has been detected in European males in England, Portugal, the Mediterranean islands of Sardinia, Italy, and Lesbos, Greece, and the Eastern Mediterranean regions of Anatolia, the Levant, and Southern Arabia.
http://en.wikipedia.org/wiki/Haplogroup_A_%28Y-DNA%29
For Haplogroup BT, it is a hypothetical grouping and no human actually has a BT Y-chromosome. Same also goes for CT and CF.
Haplogroups BT, CT, CF, are hypothetically. Until today, no male in haplogroup BT or CT or CF has yet been discovered.
http://my.opera.com/ancientmacedonia/bl … acedonians
“No male in haplogroup CT* has yet been discovered.”
http://en.wikipedia.org/wiki/Haplogroup_CT_%28Y-DNA%29
“The haplogroup is hypothetical because no male in haplogroup CF* has yet been discovered.”
http://en.wikipedia.org/wiki/Haplogroup_CF_%28Y-DNA%29
Also there is no definitive place of origin for BT either, though sources say that it probably originated in North East Africa.
Haplogroup BT split off from haplogroup A 70,000 years bp , probably originating in North East Africa from Y-chromosomal Adam. It contains all living human Y-DNA haplogroups except for A.
http://en.wikipedia.org/wiki/Haplogroup_BT_%28Y-DNA%29
The F Haplogroup contains more than 90% of the world’s population.
In human genetics, haplogroup F or FT is an enormous Y-chromosome haplogroup spanning all the continents. This haplogroup and its subclades contain more than 90% of the world’s existing male population.
http://en.wikipedia.org/wiki/Haplogroup_F_%28Y-DNA%29
It is theorized that it originated in the Middle East area.
http://www.familytreedna.com/public/F-YDNA/default.aspx
Because BT, CT, and CF are all hypothetical and not found in any human, it is entirely possible that F can be located nearer to the root of the tree. And since it accounts for 90% of the current population, this would make it seem more likely that it is close to the root.
So, though this is tentative for now, it’s possible to arrange the Y-chromosome tree into 3 descendants off of a common ancestor: AB, CF, and DE.
If the only features we could compare were morphological, there might be cause for confusion (since different sequences can sometimes encode similar structures).
Then the use of fossil evidence would be unreliable.
However, we know a mechanism that can cause the the hierarchical differences in species’ sequences.
That is what we are debating now with the concept of macroevolution.
Note that every phylogenetic tree we make assumes homology.
Would even phylogenetic trees based on genetics also assume homology?
simply show that different genes from the same set of organisms infer the same tree no more often than randomized trees.
OK, let’s spend some more time on this then.
If different genes from the same organisms point to different evolutionary pathways, these would be different trees correct?
What would constitute randomized trees?
Conversely, you have not yet described the mechanisms by which species are designed or what types of genetic data can be used to falsify this hypothesis. Maybe you will elaborate in your response to my other post.
I already addressed your post.
In terms of the mechanism that a species is designed, it not necessary to know how something is designed to posit a designer.
One example would be the SETI program. If researchers find a signal from another planet, it would not be immediately rejected because they do not know how an alien civilization created the signal.
And I did present a way for genetic data to falsify my model. But just because the bar is high doesn’t mean that I have not presented a way to falsify it using genetic data.
https://debatingchristianity.com/forum/viewtopic.php?p=324759#p324759
